Installation
BactImAs Installation Guide
1. BactImAs releases (download)
Release 0.7.1
* Binaries (ready to run) * BactImAs(6 MB) * Source code (for the developers) * Hosted at Github: https://github.com/mekterovic/bactimas
2. Installation
BactImAs is a multi-platform application written in Java which uses sqlite database. It can run (at least) on:
* Mac * Linux * Ms Windows
However, you should first install (if you don't already have it):
* Java (1.7+) [[1]] * SqLite [[2]], [guide]
both of them being easy to install.
BactImAs is distributed in two ways:
- as an [[3]] **plugin** - **stand-alone** version
If you're using the plugin, then you can easily install BactImAs via the Icy plugin infrastructure (and Icy will make sure you always have an up-to-date version).
If you want the standalone version, then the installation boils down to downloading and extracting a zip file. Once you've extracted BactImAs.zip into a folder of your choice, you start the app by:
(a) double clicking on the bactimas.jar or, from console,using the following statement (from the BactImAs folder):
(b) java -jar bactimas.jar or (c) java -jar -Xmx500m bactimas.jar (to give it e.g. 500MB of memory, depends on your configuration)
In both cases, upon start, BactImAs will detect that you don't have the data and conf folders and will offer to set them up. You should agree and subsequently choose whether you want to download the demo database (~180MB). You can also download the demo database later, either through the same process or manually.
3. BactImAs structure
BactImAs is structured as follows:
(bactimas folder)
|
|- bactimas.jar (main file, double click it to run BactImAs, or start it from the console with: java -jar BactImAs.jar)
|- IJ_Prefs.txt (IJ prefs file) - only in standalone version
|- ImageJ.cfg (IJ config file - used on Windows OS to control and save how much memory will ImageJ get, open it with any text editor and check) - only in standalone version
|- logs (logs folder, used for troubleshooting)
|- lib (folder with additional library files)
|- conf (folder with configuration files, here you specify the database and additional algorithms)
|- data (IMPORTANT: folder holding your data: database and image files.
|- images You should backup this folder periodically. )
|- dbs
4. Architecture
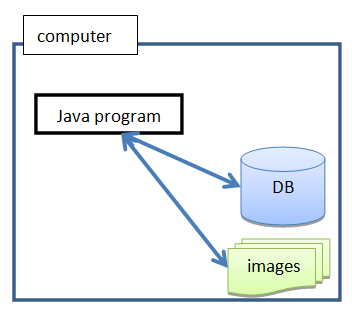
BactImAs is designed to run in a single-user mode, meaning that the program is self-sufficient and does not require internet connection to work. This is achieved by keeping the program and data (database and images) locally:
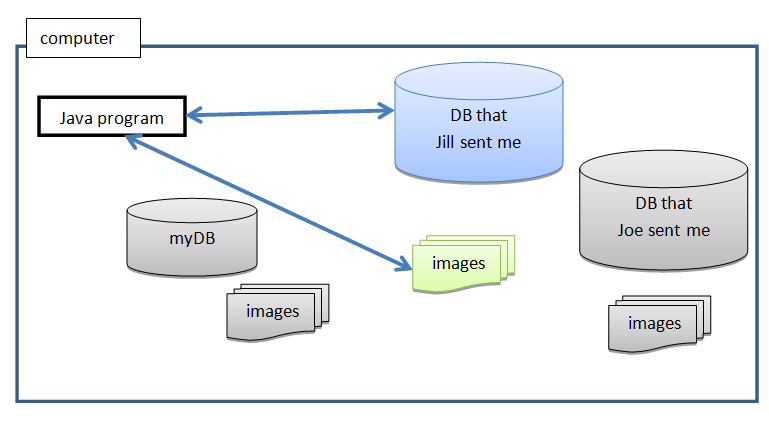
This doesn't mean that you cannot share your data with someone else. It is possible to have multiple databases on your machine (but work with only one at the time), e.g.:
Merging databases is not yet supported, but it will probably be supported in the future.
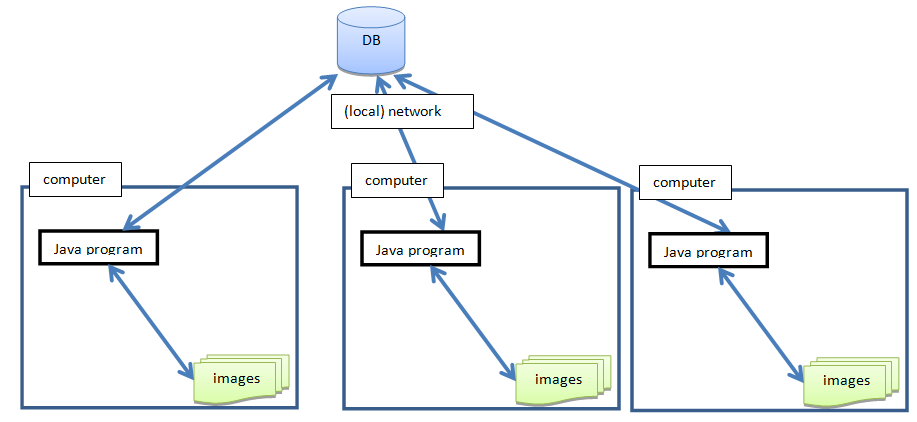
BactImAs could also be configured to run in an Intranet environment, where all users would save the data into the same database (but then it would be better to swap SQLite for some other database, e.g. MySql, PostgreSQL, etc.) maintained by the DB Administrator, e.g.:
5. Version upgrade
To upgrade, download the latest version and: (a) extract new BactImAs version into a new folder (b) copy the old version's data folder over(write) the new version's data folder.
6. Troubleshooting:
BactImAs will not start
Check which version of Java you have installed because BactImAs runs on Java 7 or higher.
* In Mac OS: Go to system preferences. In the folder “Other” doubleclick on Java icon and a new window will appear where you can click “About” to check which Java version you have installed. * In Windows, Linux, Mac: start the console (win_key+R, then type "cmd") and type "java -version" in the console to find your java version
There are no available experiments to load
If you haven't installed the empty version (in which case this is fine) then it is probably a path issue. Check whether the URL in the conf/db.properties points to the existing file. Make sure the database path is correct and you used the correct delimiter (there can be differences between OS: slash vs backslash).