Main Page: Difference between revisions
No edit summary |
No edit summary |
||
| Line 35: | Line 35: | ||
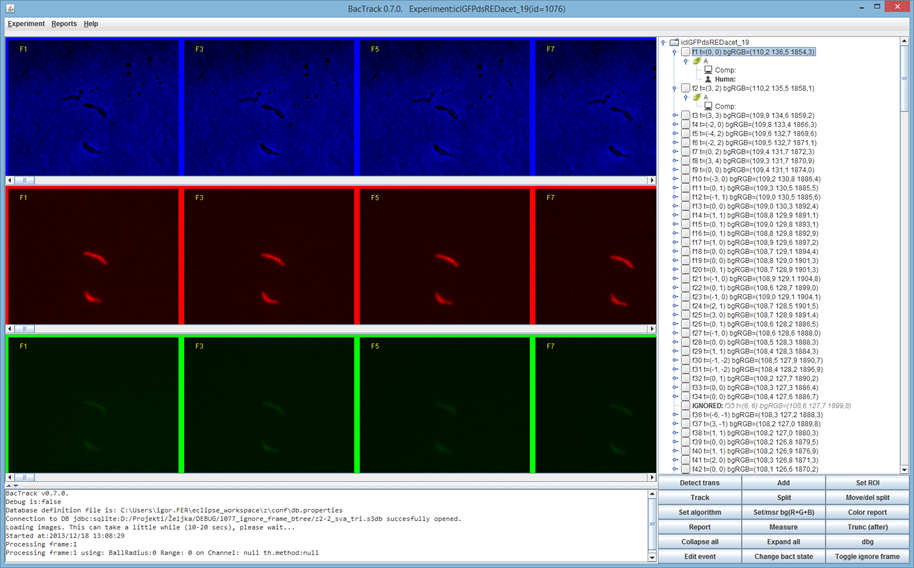
[[File:bactrack_main.png|400px]] | |||
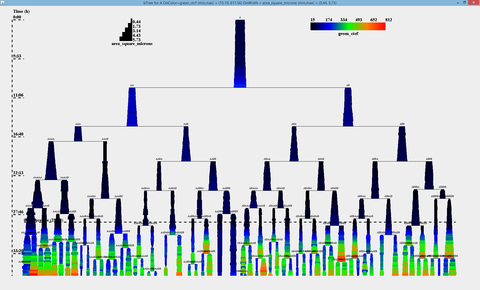
[[File:btree.png|400px]] | |||
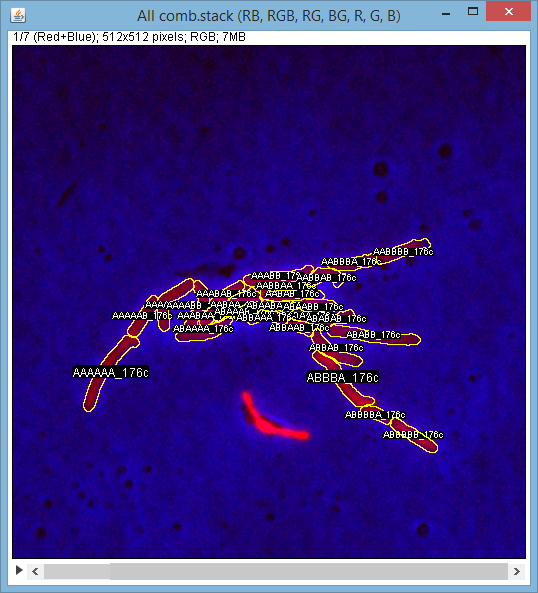
[[File:frame.png|400px]] | |||
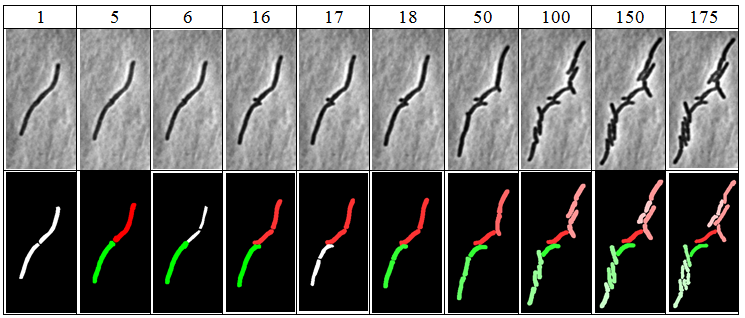
[[File:segmentation.png|400px]] | |||
| Line 47: | Line 48: | ||
---- | ---- | ||
Example reports produced by BactImAs: | Example reports produced by BactImAs: | ||
<embedvideo service="youtube">https://www.youtube.com/watch?v=xSpLbs5pe_Y</embedvideo> | |||
{{youtube>xSpLbs5pe_Y?medium|BW Report video generated from the program (black outline - computer, white - human) }} | {{youtube>xSpLbs5pe_Y?medium|BW Report video generated from the program (black outline - computer, white - human) }} | ||
{{youtube>GKMdunbu6CM?medium|Color report video generated from the program (green outline - human, red - human) }} | {{youtube>GKMdunbu6CM?medium|Color report video generated from the program (green outline - human, red - human) }} | ||
Revision as of 10:35, 18 July 2023
Welcome to BactImAs Wiki pages
BactImAs is an open-source multi platform java application intended to assist the researcher in tracking various organisms in sequences of images and obtaining and visualizing their quantitative data.
Bactimas paper is published here: [[1]]
Motivation: A common task in today’s synthetic and systems biology studies is the analysis of various features of organisms using movies obtained through time-lapse microscopy. \\ This process consists of identifying organisms on a large number of images and measuring their features whilst keeping track of their lineage which is **a very cumbersome and error prone task** for the researcher. To facilitate the analysis, various mostly automated tools and algorithms were developed in recent years but all of them targeting rather specific organisms and/or laboratory setups, since a general purpose automated solution to this detection problem is extremely hard if not impossible.
Solution: We've designed BactImAs as a "platform", that is as a collection of somewhat independent well defined modules working together. Such setup facilitates maintenance, customization and upgrades. This property is particularly useful for the tracking algorithms - in BactImAs it is possible to include multiple tracking algorithms ("best-of-breed approach") and thus use the best fitted algoritm for the problem at hand. \\ It is even possible to mix-and-match algorithms in a way to segment one part of the movie with algorithm A and the other part with algorithm B.
BactImAs uses a semi-automated approach: it relies on the user to define the initial cells and all cell divisions. Other than that, the application tracks cells using a our newly developed algorithm. At any point, the user can intervene and correct any unsatisfactory cell detections.
BactImAs provides the following features:
* Easy to use GUI interface * Frame anlignment algorithm * Novel mycobacteria segmentation algorithm * Novel configurable lineage tree visualization * Automated acquisition of quantitative date * Relational database storage, SQL interface to data * Data export (CSV)
Example reports produced by BactImAs:
<embedvideo service="youtube">https://www.youtube.com/watch?v=xSpLbs5pe_Y</embedvideo>
{{youtube>xSpLbs5pe_Y?medium|BW Report video generated from the program (black outline - computer, white - human) }} {{youtube>GKMdunbu6CM?medium|Color report video generated from the program (green outline - human, red - human) }}